Data Platform Design for Healthcare Research
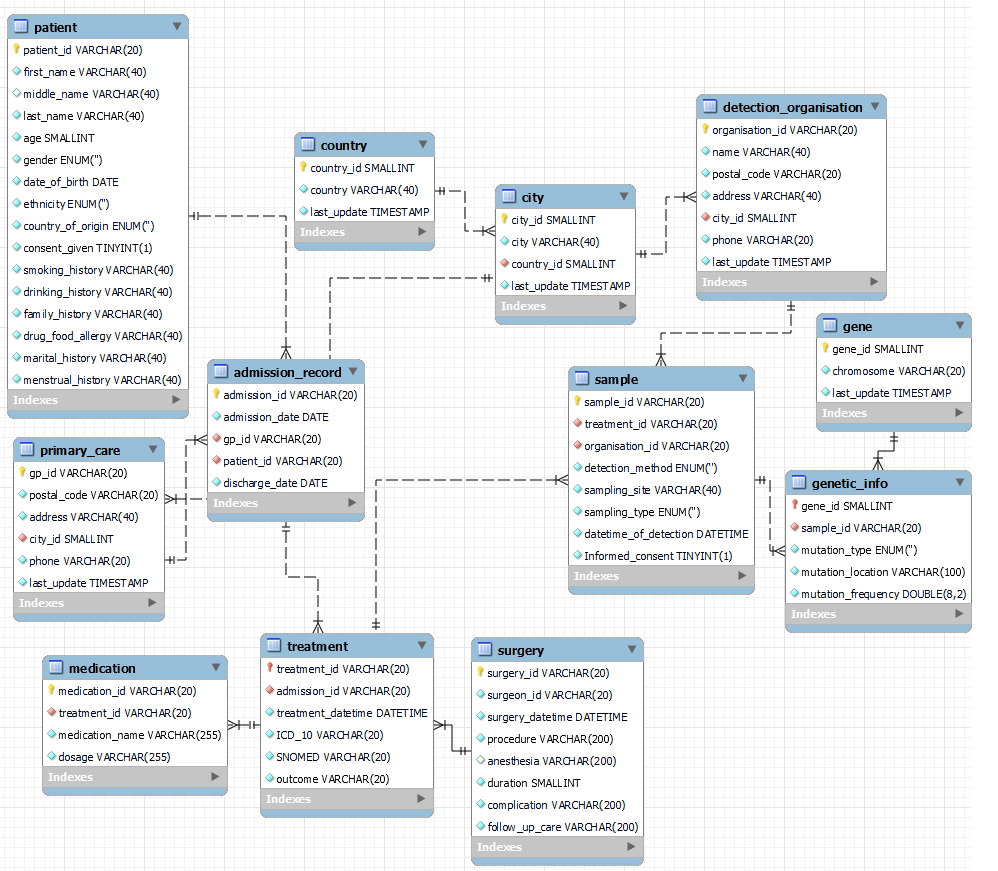
Table of Content
Chapter 1 - Project Overview
Chapter 2 - Design of Schema
Chapter 3 - Data Privacy and Security
Chapter 4 - Extract, Transform, and Load (ETL)
Chapter 5 - Monitor and Maintain
Chapter 6 - Future Work
Chapter 1 Project Overview
The project is inspired by a data engineer job description posted by Our Future Health, to identify how diseases begin and progress in people from different backgrounds. They would like to harness genetic and clinical data to create a more detailed picture of people’s health.
MySQL is a relational database which stores structured data (each row represents one record with different features), it can create several tables with relationships that connect them together, and the purpose of storing data is to reduce duplicate data and retrieve data conveniently.
There are several objectives:
Integration of genetics and healthcare data
Easily monitor and maintain in the future work
Predicted outcome: research-ready, well-curated and well-documented data
Potential audience:
Researchers (bioinformatics scientists, surgeons, oncologists, etc)
Managers in NHS
Public
Funding Organisations
Government Health Sectors
Potential stakeholders
IT (database establishment and maintenance)
Public (disease trend)
Researchers (needed data)
Chapter 2 Design of Schema
Step 1: Initial mindmap
Tables in the schema are divided into two parts:
Clinical data
Genetic data
As the mindmap shows, clinical data comes from NHS and genetic data comes from genetic detection organisations. The clinical data involves patient general information, and admission and treatment records. The genetic data involves detailed mutated genes and frequency and other parameters.
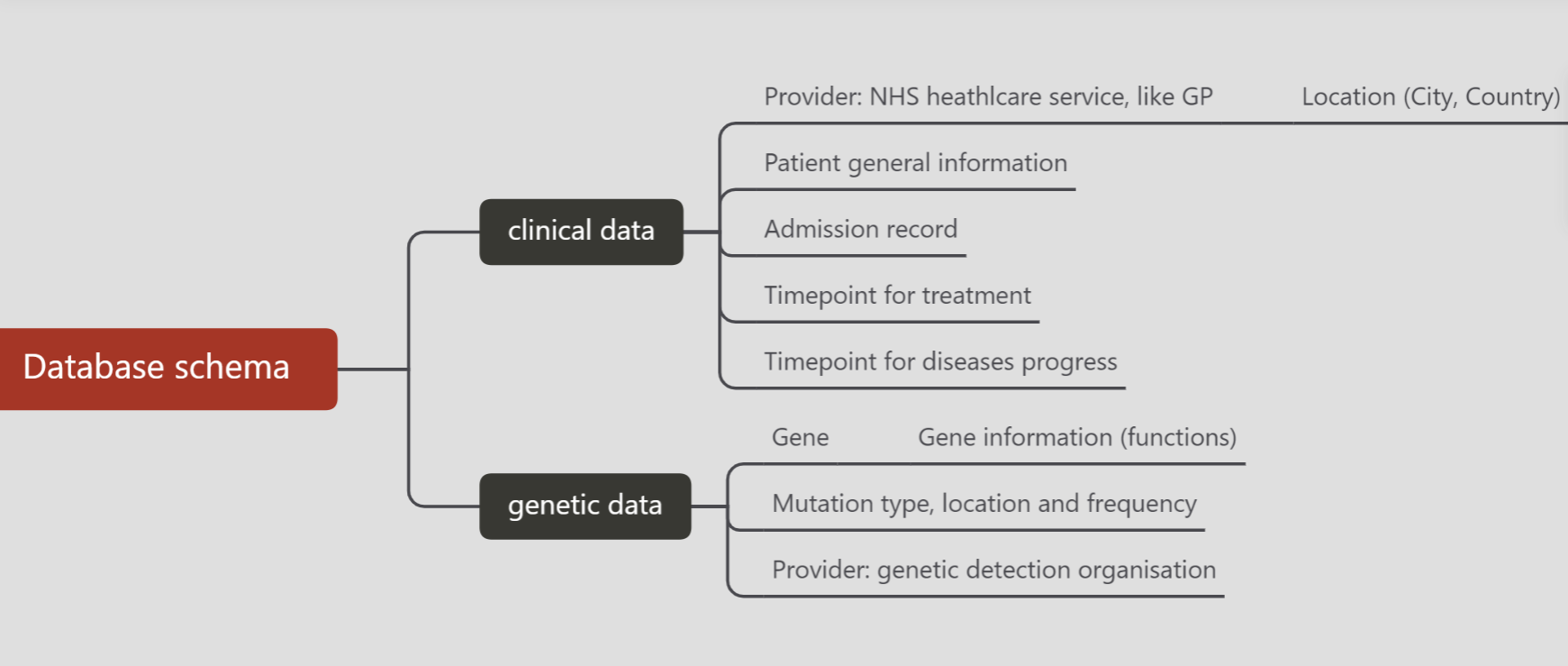
Step 2: Use drawSQL to map it
8 Tables for clinical data:
Patient
Primary care
City
Country
Admission record
Treatment
Medication
Surgery
4 Tables for genetic data:
Sample
Detection Organisation
Genetic Info
Gene
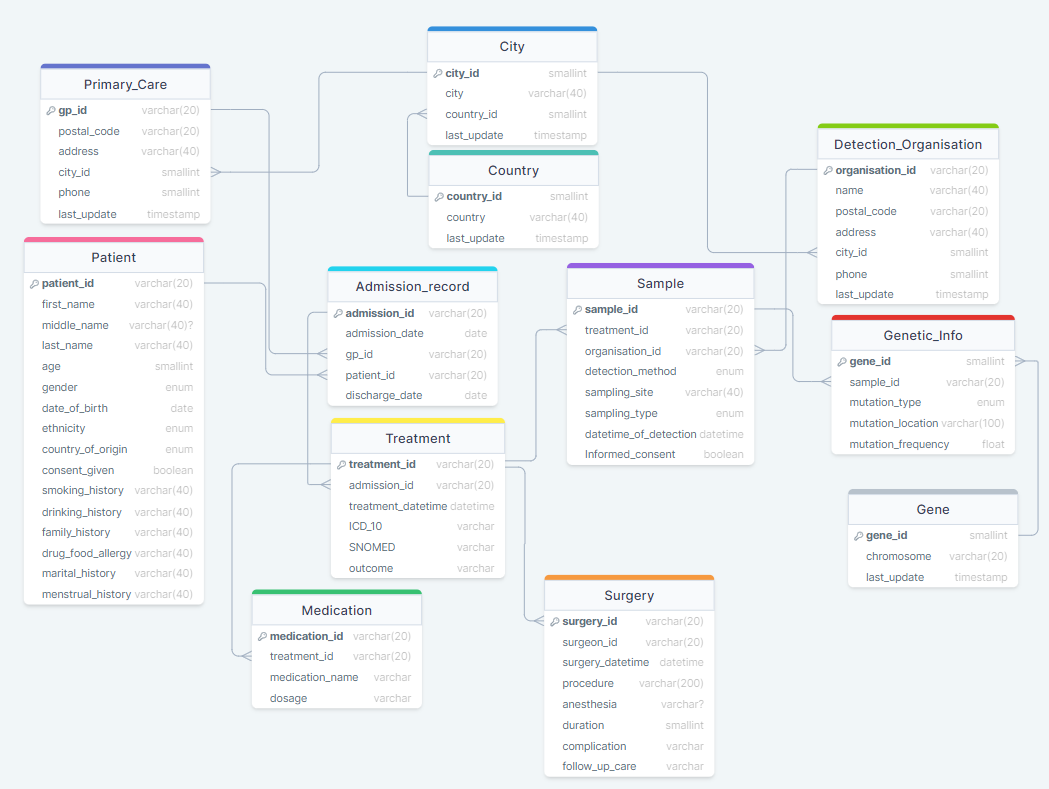
Step 3: Import SQL into MySQL
I use drawSQL to generate SQL queries, and then use MySQL to run them.
Chapter 3 Data Privacy and Security
The preparation of the database has finished, before populating the database, it is essential to talk about ethical considerations as the data is connected to patient privacy. Patients may suffer potential harm when their data is used for research purposes like revealing sensitive information. NHS currently has information governance which protects patients. "Secondary use" must only use data that will not identify individuals.
The pre-requisite includes:
Obtain the appropriate consent from patients
de-identify the data to protect patient privacy
ensure data is only used for approved purposes
Relevant technology includes:
Data anonymisation
Data encryption
Data access control
Data masking
so on
Chapter 4 Extract, Transform, and Load (ETL)
The data comes from two sources: Electronic Health Records (EHRs) and genetic detection organisations. There are several steps to do it:
obtain permission from administrators and patients
establish the relationships between EHR and the database (needed data)
extract data from EHRs (APIs, SQL)
transform: revise the message formats and map coding systems (Apache Spark)
load: import data and ensure the data is complete and accurate
Chapter 5 Monitor and Maintain
When data is generated with time and simulated in the database, the ETL process should be implemented periodically. Set up scheduled ingestion of data from the EHR databases to an analytical database (Airflow)
at the correct time
with a specific interval
in the right order
Chapter 6 Future Work
Coming back to the original purposes, the database is for healthcare researchers, considering the security and management of the database, maybe it is a good idea to set up a separate database for data analysis or views in MySQL for researchers.
Last but not least, the update for the database is an iterated process. Getting feedback from researchers or NHS managers, and adding or deleting columns will be followed in the future.
The slides I presented here. Feel free to comment on it.